Skin assessment is a test case for the CIMI architecture.
The content is based on the Pressure Ulcer Prevention Domain Analysis model and skin assessment analysis conducted by Intermountain, the US Veterans Administration, and Kaiser Permanente before and after the construction of that DAM.
Key archetypes for review include
and their ancestors and compositional inclusions.
Both Domain Analysis Models (DAMs) and CIMI models aim to capture clinical domain requirements in a format that is agnostic with respect to implementation technologies. Such specifications would support translation of information among different technologies, “future-proofing” analysis work from technological change, and confirmation that specification efforts meet commonly understood requirements.
The HL7 Development Framework describes the DAM thus:
Domain Analysis produces a set of artifacts that clearly describe the healthcare business in a given domain in terms familiar to the people who work in that business area. This set of artifacts comprises a Domain Analysis Model (DAM). HL7 workgroups use these artifacts to develop V3 standard specifications. Each artifact in the DAM must be unambiguously stated in a way that can be well understood both by the domain experts and by the HL7 project members who are responsible for developing the specification.
The CIMI project aims to produce “detailed clinical models,” i.e., models that provide the level of detail necessary for clinical use, including specific semantics. Or, as one CIMI presentation puts it,
“Create a shared repository of detailed clinical information models
These artifacts differ in two key ways:
Moving from a DAM to CIMI, then, will involve the following activities:
We decided to test the CIMI process by modeling the Skin Breakdown Risk Assessment DAM (a.k.a., Pressure Ulcer DAM). The DAM contained information delegated to other, unfinished domains, including devices, medications and conditions, and it was clear that we had neither time nor authority to complete that work. Those areas were moved out of scope. There was, however, interest in a general skin & wound assessment, for which we did have the expertise, so the work was scoped to support a skin assessment artifact. This artifact would include general skin condition information as well as a fairly complex representation of wounds.
Most of the semantics already exist in LOINC and SNOMED CT, and most of the gaps were already in process under the guidance of the subject matter experts. The remainder are being requested in SOLOR – a system designed to extend SNOMED CT and harmonize it with RxNorm and LOINC.
While CIMI models resemble DAMs in documenting clinical requirements in a way that is not specific to an implementation technology, they tend to be more precise than DAMs by providing for its properties semantic specifications that are detailed enough to support clinical use without further expert modification.
The first implication of this need is that where a DAM may present a small number of general class model diagrams, CIMI will contain a large number of specialized models. E.g., a DAM can represent the concept of a laboratory test with properties such as test kind, value, method, and reference range, but the CIMI equivalent will specify thousands of detailed models that specify, for each given test kind, what values, methods, and range types are permissible.
The second implication is semantic. Some DAMs assert semantics purely through natural language descriptions, and some endeavor to provide standard code system values to disambiguate their ideas, but CIMI models require standard codes for disambiguation. While many properties of CIMI models have no corresponding values in code systems at this point, and some may never, the goal is to specify the semantics as completely as possible.
One challenge with this approach is the richness of semantic sources. Semantics can be specified by different systems, and they can also be specified by information model documentation. Because of these multiple potential sources for meaning, with multiple origins and source team needs, it is possible for them to overlap, and therefore to conflict. E.g., if a lab test code specifies a method, but the information model also has a slot for method, then the method semantics are overdetermined, leading to redundancy and potential contradiction.
There is no way to prevent a modeler from introducing a redundant element, but we can specify a semantic framework against which such problems may be more reliably detected. The CIMI semantic framework is the SNOMED CT concept model. Adoption of this model provides a set of shared assumptions about how concepts relate and what they are called, so that if a redundant element is created, the community has the tools to identify it in a common language.
Note that there will be cases where overlap is needed, as with the method example, because different cases will expect the content differently. For instance, it may be decided that lab tests should not precoordinate methods, and that the method field in the information model should be used, but there may be cases or populations where a set of precoordinated lab test codes must be supported. The framework in these cases can stipulate that the two forms are “iso-semantic,” managing the overlap by specifying the equivalence while prohibiting the use of the overlapping techniques for specifying method in any single instance.
Note that the CIMI models do not only bind value sets, but element semantics. That is, a blood pressure observation may have a value set of body sites where the cuff is typically placed, but the semantics of the “body site” element are usually assumed to be obvious from the natural language label. But in order to create explicit semantic expressions, we need to stipulate that the data element itself has semantics, so we provide “model bindings” to assert semantics for elements (“finding site,” in this case).
We also recognize that the SNOMED CT concept model may not support CIMI requirements in all cases. Some elements may never be appropriate for representation in description logic (DL), others may be perfectly appropriate but not yet modeled, and some may fall into a debatable middle ground. For the second case, CIMI has secured access to an extension for the creation of new concepts. These new concepts may or may not be promoted to the international edition of SNOMED CT, but they will be bounded by the assumptions of the concept model.
There may also be cases where new concepts will extend the concept model—i.e., create not only new content to fill concept relationships, but create new relationships. The alignment between these new relationships and existing or proposed SNOMED CT relationships will have to be carefully managed.
One way to ensure that not only the concepts defined in the CIMI extension but also the disposition of concepts in CIMI models are semantically consistent is to use the models to infer description logic expressions and to classify them in a reasoner. If the inferred DL expressions don’t classify as expected, then the model properties have been incorrectly bound to the concepts, or concepts have been incorrectly authored. We propose to test this approach with a skin assessment example.
We anticipate that inferred expressions will also serve two other purposes:
We adopt the skin color evaluation from the skin assessment DAM as an example. This is an observation not of the wavelength of light, but of any divergence from the normal (phenotypic) appearance with potential physiological ramifications, e.g., “flushed” or “pale.” (Note: discussions continue as to whether to err on the side of perceived appearance (e.g., “blue” or “yellow”) or physiological ramification (e.g., “cyanotic” or “jaundiced”
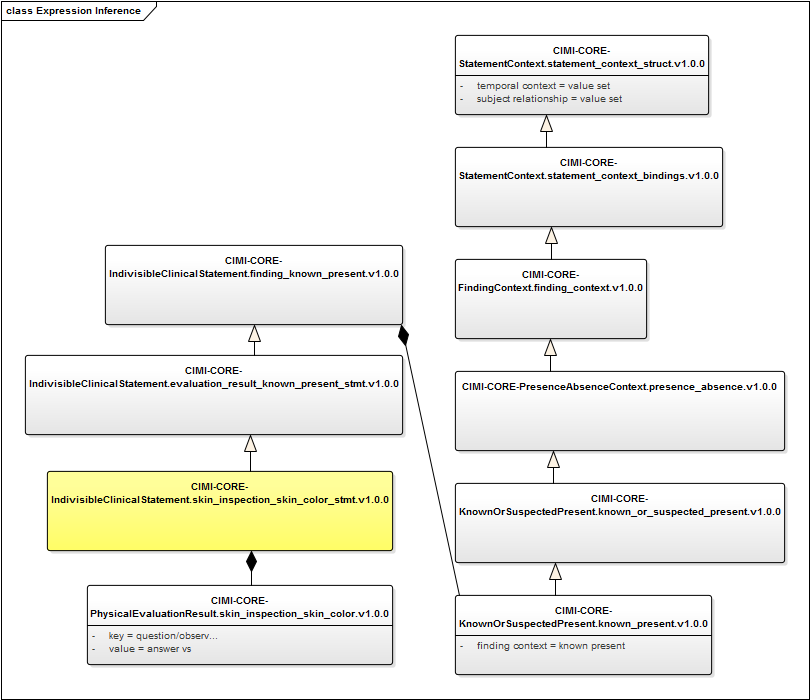
The archetype is CIMI-CORE-IndivisibleClinicalStatement.skin_inspection_skin_color_stmt.v1.0.0. Its place in the hierarchy, and the names of associated archetypes, is shown in figure 1.
Because we do have examples of the assertion of absence of phenomena elsewhere in the domain, we need to use the SNOMED CT model for Situation with Explicit Context; because we wish to be as consistent as possible, we choose to use that model even where our situation is the default one, where Finding alone would serve otherwise. (The default context for Finding is present in the patient at the current or specified time.) We recognize that this model stretches the concept of “ontology” toward that of “information model,” and we solicit specific concerns or recommendations for managing this extension.
The pseudo-code for the expected expression follows:
Note that the “Associated finding” attribute includes “observable entity” in its range.
The SNOMED CT “color of skin” concept has no definition beyond its fully specified name and its sole “is-a” relationship. (It does have a more specific child, Fitzpatrick classification, but while that ensures that a phenotypic interpretation is in scope, it does not eliminate other possibilities.) Whether this is a freedom intended by the concept’s author, an oversight, or a limitation of the authoring process, the intended semantics are not clear. The CIMI model, on the other hand, is intended to provide unambiguous semantics for the question (“divergence from the normal (phenotypic) appearance with potential physiological ramifications”), partly in terms of its range of answers.
The skin modeling group identified answers to the question, and these answers are enumerated under the LOINC code that the group authored. In LOINC, the answers are strings and not concept codes, but they provide semantic disambiguation. We faced two options for getting these values into a DL expression following the SNOMED CT concept model:
Some members expressed caution about modeling the colors as findings, as they are typically observed by nurses, and therefore cannot be considered diagnoses. However, the colors seemed reductive; “red” skin is not purely red, but only in contrast to normal skin.
The other constraint is the concept model itself. The concept model at this time has no mechanism for associating an observable entity with an evaluated result. It does, however, provide a relationship between a finding and an observable entity that the finding “interprets.” This, then, is the path we have chosen. As a result, the model appears thus:
This approach provides a place in which to put the finding values (e.g., “blue skin (finding)” or “cyanosis of skin (finding)”). However, it still fails to enumerate the acceptable values, and, in fact, DL expressions have no specific mechanism for defining value sets. SNOMED International has drafted a syntax to support value set reference, but it has no DL translation.
The approach we adopt is to define a class for DL classification in terms of its members; i.e., the value set appears in the expression as a single concept with children. It is unclear at this point whether the union concept should persist in the ontology or exist purely as a transient artifact in the DL, constructed from the results of a terminology server query. For this example, we assign the concept a temporary identifier. SNOMED CT assertions to describe this state follow:
(Note that while the answer values have been identified by clinical review, the definition of the precise semantics & encoding -- e.g., blue vs. cyanotic -- are still in process.)
This example uses nine distinct DL expressions to provide the necessary clarification of the range of the question.
Unfortunately, the open-world assumption means that this expression of the skin color parent concept does not exclude any other concepts. I.e., the above expression will include observations using the identified values, but it will not exclude observations using other values. For this, we must use the OWL “unionOf” relationship to specify the identified answer concepts disjunctively; i.e., assuming that concepts not so specified are disjoint with the value set.
Note that SNOMED conforms to EL+, which does not support disjunction (including the “unionOf” relation). This does not necessarily mean you cannot use disjunction with SNOMED values: one result of this example will be to identify any issues with the feasibility of using this approach.
The pseudocode expression in this approach would be
In order to generate an expression, each of the concepts in the expression must be found in the subject asset. In a prior incarnation of the CIMI architecture, both value set values and model binding values for element semantics were explicitly stated in the “flattened” archetype, so it was possible to generate an expression from a single file. In the current design, there is no flattened view (yet), so any attempt to parse the document for concept identifiers must be able to traverse inheritance and composition relationships to other files. In addition, concept identifiers are indicated by URIs, and these URIs identify value sets rather than individual codes, for model bindings as well as for value sets. As a result, no expression can be inferred without the support of a terminology service to resolve the value sets.
We do not provide such a utility. But we do indicate the sources of the information required to generate the expression. As the names are very long, and the relationships complex, we present the relationships in a diagram.
Figure 1: Graph of Skin Color archetypes and relationships
In this diagram, the archetype of interest is shown in yellow, and the Situation attribute values are accessible by de-referencing the composition and inheritance relationships.
First, the archetype specifies inclusion of a topic archetype. This archetype specifies “key” and “value” fields. These are equivalent to LOINC question (or SNOMED observable) and answer (or SNOMED finding) values. The archetype specifies them as URIs: the question to http://example.com/valueset/skin_color_vs.v0.0.1 and its answers to http://example.com/valueset/skin_color_range_vs.v0.0.1.
The archetype’s grandparent includes the context archetype Known or Suspected Present, which constrains the Finding Context value.
And four generations above that archetype, the context pattern establishes the Finding Context property, with its semantic binding to the SNOMED attribute, as well as the Temporal context and relationship to subject of record.
We require a service to return value sets for the URIs specified in the archetypes. Model bindings to attributes will typically be to a single code, but may be to sets (e.g., procedure device, which has four children). The formalism, however, uses the value set construct uniformly: single-code sets will need to be de-referenced as well.
Note also that some concepts (e.g., CIMI_Skin_Color_Evaluation_Result (finding)) may not be present in SNOMED. The service must use a CIMI-aware knowledge base, and it is there that the definition of that concept must be asserted, including the unionOf property and the interprets property.
An automated process can derive the expression outlined above by parsing the archetype tree.
The model itself can be validated by constructing and classifying the expression. Note that the expression will not fully represent the model, which will contain elements not represented in the expression. Equivalent concepts and near-equivalent concepts can be used to confirm the appropriate classification.
This exercise can be completed most easily on a flattened archetype, and it cannot be completed without a terminology server.